Biopax-Explorer code documentation#
biopax-explorer.biopax.dna Module#
Classes#
|
Methods |
|
Class Dna |
|
Methods |
|
Class PhysicalEntity |
Class Inheritance Diagram#

biopax-explorer.biopax Package#
biopax-explorer.graph.serializer Module#
Functions#
Generates a BioPAX RDF template. |
Classes#
|
Methods |
Class Inheritance Diagram#

biopax-explorer.graph.view Module#
Functions#
|
Checks the installed backends and returns a message indicating the available options. |
|
Generates a string representation of a graph. |
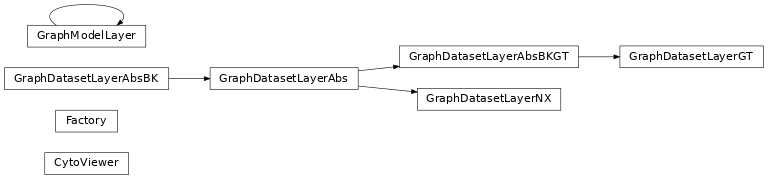
Classes#
|
A wrapper for the cytoscape.js jupyter widget. |
|
Factory class for creating graph dataset layers. |
GraphDatasetLayerGT class for Graph-tool backend. |
|
GraphDatasetLayerNX class for NetworkX backend. |
|
GraphModelLayer class. |
Class Inheritance Diagram#
biopax-explorer.query.client Module#
Classes#
|
A class for interacting with a BIOPAX RDF store. |
|
Class BindingFeature |
|
Class BioSource |
|
Class BiochemicalPathwayStep |
|
Class BiochemicalReaction |
|
Class Catalysis |
|
Class CellVocabulary |
|
Class CellularLocationVocabulary |
|
Class ChemicalStructure |
|
Class Complex |
|
Class ComplexAssembly |
|
Class Control |
|
Class ControlledVocabulary |
|
Class Conversion |
|
Methods |
|
Xml and Schema data types to native python. |
|
Class Degradation |
|
Class DeltaG |
|
Class Dna |
|
Class DnaReference |
|
Class DnaRegion |
|
Class DnaRegionReference |
|
Class Entity |
|
Class EntityFeature |
|
Class EntityReference |
|
Class EntityReferenceTypeVocabulary |
|
The core component of Jinja is the Environment. |
|
Class Evidence |
|
Class EvidenceCodeVocabulary |
|
Class ExperimentalForm |
|
Class ExperimentalFormVocabulary |
|
Load templates from a directory in the file system. |
|
Class FragmentFeature |
|
Class Gene |
|
Class GeneticInteraction |
|
An RDF Graph |
|
Methods |
|
Methods |
|
Methods |
|
Methods |
|
Class Interaction |
|
Class InteractionVocabulary |
|
Class KPrime |
|
RDF 1.1's Literals Section: http://www.w3.org/TR/rdf-concepts/#section-Graph-Literal |
|
Methods |
|
Methods |
|
Methods |
|
Methods |
|
Class ModificationFeature |
|
Class Modulation |
|
Class MolecularInteraction |
|
Utility class for quickly generating URIRefs with a common prefix |
|
PurePath subclass that can make system calls. |
|
Class Pathway |
|
Class PathwayStep |
|
Class PhenotypeVocabulary |
|
Class PhysicalEntity |
|
Class Protein |
|
Class ProteinReference |
|
Class Provenance |
|
Class PublicationXref |
|
Methods |
|
Common qualified names. |
|
The RDF Concepts Vocabulary (RDF) |
|
Class RelationshipTypeVocabulary |
|
Class RelationshipXref |
|
Class Rna |
|
Class RnaReference |
|
Class RnaRegion |
|
Class RnaRegionReference |
|
Wrapper around an online access to a SPARQL Web entry point. |
|
Class Score |
|
Class SequenceInterval |
|
Class SequenceLocation |
|
Class SequenceModificationVocabulary |
|
Class SequenceRegionVocabulary |
|
Class SequenceSite |
|
Class SmallMolecule |
|
Class SmallMoleculeReference |
|
Class Stoichiometry |
|
Methods |
|
Methods |
|
Methods |
|
Class TemplateReaction |
|
Class TemplateReactionRegulation |
|
Class TissueVocabulary |
|
Class Transport |
|
Methods |
|
RDF 1.1's IRI Section https://www.w3.org/TR/rdf11-concepts/#section-IRIs |
|
Class UnificationXref |
|
Class UtilityClass |
|
Class Xref |
|
islice(iterable, stop) --> islice object islice(iterable, start, stop[, step]) --> islice object |
Class Inheritance Diagram#

biopax-explorer.pattern.rack Module#
Functions#
|
Classes#
|
Class BindingFeature |
|
Class BioSource |
|
Class BiochemicalPathwayStep |
|
Class BiochemicalReaction |
|
Class Catalysis |
|
Class CellVocabulary |
|
Class CellularLocationVocabulary |
|
Class ChemicalStructure |
|
Class Complex |
|
Class ComplexAssembly |
|
Class Control |
|
Class ControlledVocabulary |
|
Class Conversion |
|
Methods |
|
Class Degradation |
|
Class DeltaG |
|
Class Dna |
|
Class DnaReference |
|
Class DnaRegion |
|
Class DnaRegionReference |
|
Class Entity |
|
Class EntityFeature |
|
Methods |
|
Class EntityReference |
|
Class EntityReferenceTypeVocabulary |
|
Class Evidence |
|
Class EvidenceCodeVocabulary |
|
Class ExperimentalForm |
|
Class ExperimentalFormVocabulary |
|
Class FragmentFeature |
|
Class Gene |
|
Class GeneticInteraction |
|
Class Interaction |
|
Class InteractionVocabulary |
|
Class KPrime |
|
LocalProcessing class represents local processing operations. |
|
Class ModificationFeature |
|
Class Modulation |
|
Class MolecularInteraction |
|
PurePath subclass that can make system calls. |
|
Class Pathway |
|
Class PathwayStep |
|
Pattern class represents a container for processing steps. |
|
Class PhenotypeVocabulary |
|
Class PhysicalEntity |
|
A class representing a processing collection. |
|
Class Protein |
|
Class ProteinReference |
|
Class Provenance |
|
Class PublicationXref |
|
A reference list of Patterns related to BIOPAX use cases. |
|
Class RelationshipTypeVocabulary |
|
Class RelationshipXref |
|
Class Rna |
|
Class RnaReference |
|
Class RnaRegion |
|
Class RnaRegionReference |
|
Class Score |
|
Class SequenceInterval |
|
Class SequenceLocation |
|
Class SequenceModificationVocabulary |
|
Class SequenceRegionVocabulary |
|
Class SequenceSite |
|
Class SmallMolecule |
|
Class SmallMoleculeReference |
|
Step class represents a step in the processing sequence of a Pattern. |
|
Class Stoichiometry |
|
Class TemplateReaction |
|
Class TemplateReactionRegulation |
|
Class TissueVocabulary |
|
Class Transport |
|
Methods |
|
Class UnificationXref |
|
Class UtilityClass |
|
Class Xref |
Class Inheritance Diagram#

biopax-explorer.pattern.processing Module#
Functions#
|
Filter the collection using a blacklist. |
|
|
|
Eliminate the matches where the small molecule at the left and at the right is the same. |
Classes#
|
Class BindingFeature |
|
Class BioSource |
|
Class BiochemicalPathwayStep |
|
Class BiochemicalReaction |
|
A class representing a Black List Filter. |
|
Class Catalysis |
|
Class CellVocabulary |
|
Class CellularLocationVocabulary |
|
Class ChemicalStructure |
|
Class Complex |
|
Class ComplexAssembly |
|
Class Control |
|
Class ControlledVocabulary |
|
Class Conversion |
|
Methods |
|
Class Degradation |
|
Class DeltaG |
|
Class Dna |
|
Class DnaReference |
|
Class DnaRegion |
|
Class DnaRegionReference |
|
Class Entity |
|
Class EntityFeature |
|
Class EntityReference |
|
Class EntityReferenceTypeVocabulary |
|
Class Evidence |
|
Class EvidenceCodeVocabulary |
|
Class ExperimentalForm |
|
Class ExperimentalFormVocabulary |
|
Class FragmentFeature |
|
Class Gene |
|
Class GeneticInteraction |
|
Class Interaction |
|
Class InteractionVocabulary |
|
Class KPrime |
|
Class ModificationFeature |
|
Class Modulation |
|
Class MolecularInteraction |
|
Methods |
|
PurePath subclass that can make system calls. |
|
Class Pathway |
|
Class PathwayStep |
|
Class PhenotypeVocabulary |
|
Class PhysicalEntity |
A class representing a processing collection. |
|
|
Class Protein |
|
Class ProteinReference |
|
Class Provenance |
|
Class PublicationXref |
|
Class RelationshipTypeVocabulary |
|
Class RelationshipXref |
|
Class Rna |
|
Class RnaReference |
|
Class RnaRegion |
|
Class RnaRegionReference |
|
Class Score |
|
Class SequenceInterval |
|
Class SequenceLocation |
|
Class SequenceModificationVocabulary |
|
Class SequenceRegionVocabulary |
|
Class SequenceSite |
|
Class SmallMolecule |
|
Class SmallMoleculeReference |
|
Class Stoichiometry |
|
Class TemplateReaction |
|
Class TemplateReactionRegulation |
|
Class TissueVocabulary |
|
Class Transport |
|
Methods |
|
Class UnificationXref |
|
Class UtilityClass |
|
Class Xref |
Class Inheritance Diagram#
biopax-explorer.pattern.view Module#
Functions#
|
Converts DOT file to PNG image. |
|
Converts DOT file to SVG image. |
|
Converts DOT string to PNG image. |
|
Converts DOT string to SVG image. |
|
Generates a graph view of a given Pattern and writes it to an image file. |
biopax-explorer.pattern.pattern Module#
Classes#
|
This class fills the entities with void attributes. |
This class manages 'in-memory processing' to post-process a SPARQL query. |
|
|
Utility class to mock model entities with a lower memory footprint. |
|
This class holds a list of Step objects containing Patterns or LocalProcessings. |
|
This class manages the execution of a 'Pattern' instance. |
This class manages a list of referenced 'in-memory processing'. |
|
|
This class holds two kinds of data: - a list of EntityNodes used to generate a SPARQL query, or - a LocalProcessing that enables local in-memory filter operations on entities. |
Class Inheritance Diagram#
biopax-explorer.biopax.bindingfeature Module#
Classes#
|
Class BindingFeature |
|
Methods |
|
Class EntityFeature |
|
Methods |
Class Inheritance Diagram#

biopax-explorer.biopax.biochemicalpathwaystep Module#
Classes#
|
Class BiochemicalPathwayStep |
|
Methods |
|
Methods |
|
Class PathwayStep |
Class Inheritance Diagram#

biopax-explorer.biopax.biochemicalreaction Module#
Classes#
|
Class BiochemicalReaction |
|
Methods |
|
Class Conversion |
|
Methods |
Class Inheritance Diagram#

biopax-explorer.biopax.biosource Module#
Classes#
|
Class BioSource |
|
Methods |
|
Methods |
|
Class UtilityClass |
Class Inheritance Diagram#

biopax-explorer.biopax.catalysis Module#
Classes#
|
Methods |
|
Class Catalysis |
|
Class Control |
|
Methods |
Class Inheritance Diagram#

biopax-explorer.biopax.cellularlocationvocabulary Module#
Classes#
|
Methods |
|
Class CellularLocationVocabulary |
|
Class ControlledVocabulary |
|
Methods |
Class Inheritance Diagram#

biopax-explorer.biopax.cellvocabulary Module#
Classes#
|
Methods |
|
Class CellVocabulary |
|
Class ControlledVocabulary |
|
Methods |
Class Inheritance Diagram#

biopax-explorer.biopax.chemicalstructure Module#
Classes#
|
Methods |
|
Class ChemicalStructure |
|
Methods |
|
Class UtilityClass |
Class Inheritance Diagram#

biopax-explorer.biopax.complex Module#
Classes#
|
Methods |
|
Class Complex |
|
Methods |
|
Class PhysicalEntity |
Class Inheritance Diagram#

biopax-explorer.biopax.complexassembly Module#
Classes#
|
Methods |
|
Class ComplexAssembly |
|
Class Conversion |
|
Methods |
Class Inheritance Diagram#

biopax-explorer.biopax.control Module#
Classes#
|
Methods |
|
Class Control |
|
Methods |
|
Class Interaction |
Class Inheritance Diagram#

biopax-explorer.biopax.controlledvocabulary Module#
Classes#
|
Methods |
|
Class ControlledVocabulary |
|
Methods |
|
Class UtilityClass |
Class Inheritance Diagram#

biopax-explorer.biopax.conversion Module#
Classes#
|
Methods |
|
Class Conversion |
|
Methods |
|
Class Interaction |
Class Inheritance Diagram#

biopax-explorer.biopax.covalentbindingfeature Module#
Classes#
|
Methods |
|
Methods |
|
Methods |
Class Inheritance Diagram#

biopax-explorer.biopax.degradation Module#
Classes#
|
Methods |
|
Class Conversion |
|
Class Degradation |
|
Methods |
Class Inheritance Diagram#

biopax-explorer.biopax.deltag Module#
Classes#
|
Methods |
|
Class DeltaG |
|
Methods |
|
Class UtilityClass |
Class Inheritance Diagram#

biopax-explorer.biopax.dna Module#
Classes#
|
Methods |
|
Class Dna |
|
Methods |
|
Class PhysicalEntity |
Class Inheritance Diagram#

biopax-explorer.biopax.dnareference Module#
Classes#
|
Methods |
|
Class DnaReference |
|
Class EntityReference |
|
Methods |
Class Inheritance Diagram#

biopax-explorer.biopax.dnaregion Module#
Classes#
|
Methods |
|
Class DnaRegion |
|
Methods |
|
Class PhysicalEntity |
Class Inheritance Diagram#

biopax-explorer.biopax.dnaregionreference Module#
Classes#
|
Methods |
|
Class DnaRegionReference |
|
Class EntityReference |
|
Methods |
Class Inheritance Diagram#

biopax-explorer.biopax.entity Module#
Classes#
|
Methods |
|
Class Entity |
|
Methods |
Class Inheritance Diagram#

biopax-explorer.biopax.entityfeature Module#
Classes#
|
Methods |
|
Class EntityFeature |
|
Methods |
|
Class UtilityClass |
Class Inheritance Diagram#

biopax-explorer.biopax.entityreference Module#
Classes#
|
Methods |
|
Class EntityReference |
|
Methods |
|
Class UtilityClass |
Class Inheritance Diagram#

biopax-explorer.biopax.entityreferencetypevocabulary Module#
Classes#
|
Methods |
|
Class ControlledVocabulary |
|
Class EntityReferenceTypeVocabulary |
|
Methods |
Class Inheritance Diagram#

biopax-explorer.biopax.evidence Module#
Classes#
|
Methods |
|
Class Evidence |
|
Methods |
Class Inheritance Diagram#

biopax-explorer.biopax.evidencecodevocabulary Module#
Classes#
|
Methods |
|
Class ControlledVocabulary |
|
Class EvidenceCodeVocabulary |
|
Methods |
Class Inheritance Diagram#

biopax-explorer.biopax.experimentalform Module#
Classes#
|
Methods |
|
Class ExperimentalForm |
|
Methods |
|
Class UtilityClass |
Class Inheritance Diagram#

biopax-explorer.biopax.experimentalformvocabulary Module#
Classes#
|
Methods |
|
Class ControlledVocabulary |
|
Class ExperimentalFormVocabulary |
|
Methods |
Class Inheritance Diagram#

biopax-explorer.biopax.fragmentfeature Module#
Classes#
|
Methods |
|
Class EntityFeature |
|
Class FragmentFeature |
|
Methods |
Class Inheritance Diagram#

biopax-explorer.biopax.gene Module#
Classes#
|
Methods |
|
Class Entity |
|
Methods |
|
Class Gene |
Class Inheritance Diagram#

biopax-explorer.biopax.geneticinteraction Module#
Classes#
|
Methods |
|
Methods |
|
Class GeneticInteraction |
|
Class Interaction |
Class Inheritance Diagram#

biopax-explorer.biopax.interaction Module#
Classes#
|
Methods |
|
Class Entity |
|
Methods |
|
Class Interaction |
Class Inheritance Diagram#

biopax-explorer.biopax.interactionvocabulary Module#
Classes#
|
Methods |
|
Class ControlledVocabulary |
|
Methods |
|
Class InteractionVocabulary |
Class Inheritance Diagram#

biopax-explorer.biopax.kprime Module#
Classes#
|
Methods |
|
Methods |
|
Class KPrime |
|
Class UtilityClass |
Class Inheritance Diagram#

biopax-explorer.biopax.modificationfeature Module#
Classes#
|
Methods |
|
Class EntityFeature |
|
Methods |
|
Class ModificationFeature |
Class Inheritance Diagram#

biopax-explorer.biopax.modulation Module#
Classes#
|
Methods |
|
Class Control |
|
Methods |
|
Class Modulation |
Class Inheritance Diagram#

biopax-explorer.biopax.molecularinteraction Module#
Classes#
|
Methods |
|
Methods |
|
Class Interaction |
|
Class MolecularInteraction |
Class Inheritance Diagram#

biopax-explorer.biopax.pathway Module#
Classes#
|
Methods |
|
Class Entity |
|
Methods |
|
Class Pathway |
Class Inheritance Diagram#

biopax-explorer.biopax.pathwaystep Module#
Classes#
|
Methods |
|
Methods |
|
Class PathwayStep |
|
Class UtilityClass |
Class Inheritance Diagram#

biopax-explorer.biopax.phenotypevocabulary Module#
Classes#
|
Methods |
|
Class ControlledVocabulary |
|
Methods |
|
Class PhenotypeVocabulary |
Class Inheritance Diagram#

biopax-explorer.biopax.physicalentity Module#
Classes#
|
Methods |
|
Class Entity |
|
Methods |
|
Class PhysicalEntity |
Class Inheritance Diagram#

biopax-explorer.biopax.protein Module#
Classes#
|
Methods |
|
Methods |
|
Class PhysicalEntity |
|
Class Protein |
Class Inheritance Diagram#

biopax-explorer.biopax.proteinreference Module#
Classes#
|
Methods |
|
Class EntityReference |
|
Methods |
|
Class ProteinReference |
Class Inheritance Diagram#

biopax-explorer.biopax.provenance Module#
Classes#
|
Methods |
|
Methods |
|
Class Provenance |
|
Class UtilityClass |
Class Inheritance Diagram#

biopax-explorer.biopax.publicationxref Module#
Classes#
|
Methods |
|
Methods |
|
Class PublicationXref |
|
Class Xref |
Class Inheritance Diagram#

biopax-explorer.biopax.relationshiptypevocabulary Module#
Classes#
|
Methods |
|
Class ControlledVocabulary |
|
Methods |
|
Class RelationshipTypeVocabulary |
Class Inheritance Diagram#

biopax-explorer.biopax.relationshipxref Module#
Classes#
|
Methods |
|
Methods |
|
Class RelationshipXref |
|
Class Xref |
Class Inheritance Diagram#

biopax-explorer.biopax.rna Module#
Classes#
|
Methods |
|
Methods |
|
Class PhysicalEntity |
|
Class Rna |
Class Inheritance Diagram#

biopax-explorer.biopax.rnareference Module#
Classes#
|
Methods |
|
Class EntityReference |
|
Methods |
|
Class RnaReference |
Class Inheritance Diagram#

biopax-explorer.biopax.rnaregion Module#
Classes#
|
Methods |
|
Methods |
|
Class PhysicalEntity |
|
Class RnaRegion |
Class Inheritance Diagram#

biopax-explorer.biopax.rnaregionreference Module#
Classes#
|
Methods |
|
Class EntityReference |
|
Methods |
|
Class RnaRegionReference |
Class Inheritance Diagram#

biopax-explorer.biopax.score Module#
Classes#
|
Methods |
|
Methods |
|
Class Score |
|
Class UtilityClass |
Class Inheritance Diagram#

biopax-explorer.biopax.sequenceinterval Module#
Classes#
|
Methods |
|
Methods |
|
Class SequenceInterval |
|
Class SequenceLocation |
Class Inheritance Diagram#

biopax-explorer.biopax.sequencelocation Module#
Classes#
|
Methods |
|
Methods |
|
Class SequenceLocation |
|
Class UtilityClass |
Class Inheritance Diagram#

biopax-explorer.biopax.sequencemodificationvocabulary Module#
Classes#
|
Methods |
|
Class ControlledVocabulary |
|
Methods |
|
Class SequenceModificationVocabulary |
Class Inheritance Diagram#

biopax-explorer.biopax.sequenceregionvocabulary Module#
Classes#
|
Methods |
|
Class ControlledVocabulary |
|
Methods |
|
Class SequenceRegionVocabulary |
Class Inheritance Diagram#

biopax-explorer.biopax.sequencesite Module#
Classes#
|
Methods |
|
Methods |
|
Class SequenceLocation |
|
Class SequenceSite |
Class Inheritance Diagram#

biopax-explorer.biopax.smallmolecule Module#
Classes#
|
Methods |
|
Methods |
|
Class PhysicalEntity |
|
Class SmallMolecule |
Class Inheritance Diagram#

biopax-explorer.biopax.smallmoleculereference Module#
Classes#
|
Methods |
|
Class EntityReference |
|
Methods |
|
Class SmallMoleculeReference |
Class Inheritance Diagram#

biopax-explorer.biopax.stoichiometry Module#
Classes#
|
Methods |
|
Methods |
|
Class Stoichiometry |
|
Class UtilityClass |
Class Inheritance Diagram#

biopax-explorer.biopax.templatereaction Module#
Classes#
|
Methods |
|
Methods |
|
Class Interaction |
|
Class TemplateReaction |
Class Inheritance Diagram#

biopax-explorer.biopax.templatereactionregulation Module#
Classes#
|
Methods |
|
Class Control |
|
Methods |
|
Class TemplateReactionRegulation |
Class Inheritance Diagram#

biopax-explorer.biopax.tissuevocabulary Module#
Classes#
|
Methods |
|
Class ControlledVocabulary |
|
Methods |
|
Class TissueVocabulary |
Class Inheritance Diagram#

biopax-explorer.biopax.transport Module#
Classes#
|
Methods |
|
Class Conversion |
|
Methods |
|
Class Transport |
Class Inheritance Diagram#

biopax-explorer.biopax.transportwithbiochemicalreaction Module#
Classes#
|
Methods |
|
Methods |
|
Methods |
Class Inheritance Diagram#

biopax-explorer.biopax.unificationxref Module#
Classes#
|
Methods |
|
Methods |
|
Class UnificationXref |
|
Class Xref |
Class Inheritance Diagram#

biopax-explorer.biopax.utilityclass Module#
Classes#
|
Methods |
|
Methods |
|
Class UtilityClass |
Class Inheritance Diagram#

biopax-explorer.biopax.xref Module#
Classes#
|
Methods |
|
Methods |
|
Class UtilityClass |
|
Class Xref |
Class Inheritance Diagram#

biopax-explorer.biopax.utils.class_utils Module#
Functions#
Classes#
|
CustomEncoder : a Custom Class for instance serialization |
|
A Class to limit recursion in serialization |
|
Construct a new Decimal object. |
Class Inheritance Diagram#

biopax-explorer.biopax.utils.gen_utils Module#
Functions#
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
Classes#
|
Class BindingFeature |
|
Class BioSource |
|
Class BiochemicalPathwayStep |
|
Class BiochemicalReaction |
|
Class Catalysis |
|
Class CellVocabulary |
|
Class CellularLocationVocabulary |
|
Class ChemicalStructure |
|
Class Complex |
|
Class ComplexAssembly |
|
Class Control |
|
Class ControlledVocabulary |
|
Class Conversion |
|
Methods |
|
Class Degradation |
|
Class DeltaG |
|
Class Dna |
|
Class DnaReference |
|
Class DnaRegion |
|
Class DnaRegionReference |
|
Class Entity |
|
Class EntityFeature |
|
Class EntityReference |
|
Class EntityReferenceTypeVocabulary |
|
Class Evidence |
|
Class EvidenceCodeVocabulary |
|
Class ExperimentalForm |
|
Class ExperimentalFormVocabulary |
|
Class FragmentFeature |
|
Class Gene |
|
Class GeneticInteraction |
|
Class Interaction |
|
Class InteractionVocabulary |
|
Class KPrime |
|
Methods |
|
Class ModificationFeature |
|
Class Modulation |
|
Class MolecularInteraction |
Methods |
|
|
Class Pathway |
|
Class PathwayStep |
|
Class PhenotypeVocabulary |
|
Class PhysicalEntity |
|
Class Protein |
|
Class ProteinReference |
|
Class Provenance |
|
Class PublicationXref |
|
Class RelationshipTypeVocabulary |
|
Class RelationshipXref |
|
Class Rna |
|
Class RnaReference |
|
Class RnaRegion |
|
Class RnaRegionReference |
|
Class Score |
|
Class SequenceInterval |
|
Class SequenceLocation |
|
Class SequenceModificationVocabulary |
|
Class SequenceRegionVocabulary |
|
Class SequenceSite |
|
Class SmallMolecule |
|
Class SmallMoleculeReference |
|
Class Stoichiometry |
|
Class TemplateReaction |
|
Class TemplateReactionRegulation |
|
Class TissueVocabulary |
|
Class Transport |
|
Methods |
|
Class UnificationXref |
|
Class UtilityClass |
|
Class Xref |
Class Inheritance Diagram#

biopax-explorer.biopax.utils.validate_utils Module#
Functions#
|
|
|
|
|
|
|
|
|
Decorator factory to apply update_wrapper() to a wrapper function |
Classes#
|
|
|
|
|
Methods |
|
|
|
Methods |
|
|
|
|
|
|
|
Exception raised when failed to convert data. |
|
Methods |
Class Inheritance Diagram#

biopax-explorer.biopax.doc.dh_bindingfeature Module#
Classes#
Class bindingfeature_DocHelper |
Class Inheritance Diagram#

biopax-explorer.biopax.doc.dh_biochemicalpathwaystep Module#
Classes#
Class biochemicalpathwaystep_DocHelper |
Class Inheritance Diagram#

biopax-explorer.biopax.doc.dh_biochemicalreaction Module#
Classes#
Class biochemicalreaction_DocHelper |
Class Inheritance Diagram#

biopax-explorer.biopax.doc.dh_biosource Module#
Classes#
Class biosource_DocHelper |
Class Inheritance Diagram#

biopax-explorer.biopax.doc.dh_catalysis Module#
Classes#
Class catalysis_DocHelper |
Class Inheritance Diagram#

biopax-explorer.biopax.doc.dh_cellularlocationvocabulary Module#
Classes#
Class cellularlocationvocabulary_DocHelper |
Class Inheritance Diagram#

biopax-explorer.biopax.doc.dh_cellvocabulary Module#
Classes#
Class cellvocabulary_DocHelper |
Class Inheritance Diagram#

biopax-explorer.biopax.doc.dh_chemicalstructure Module#
Classes#
Class chemicalstructure_DocHelper |
Class Inheritance Diagram#

biopax-explorer.biopax.doc.dh_complex Module#
Classes#
Class complex_DocHelper |
Class Inheritance Diagram#

biopax-explorer.biopax.doc.dh_complexassembly Module#
Classes#
Class complexassembly_DocHelper |
Class Inheritance Diagram#

biopax-explorer.biopax.doc.dh_control Module#
Classes#
Class control_DocHelper |
Class Inheritance Diagram#

biopax-explorer.biopax.doc.dh_controlledvocabulary Module#
Classes#
Class controlledvocabulary_DocHelper |
Class Inheritance Diagram#

biopax-explorer.biopax.doc.dh_conversion Module#
Classes#
Class conversion_DocHelper |
Class Inheritance Diagram#

biopax-explorer.biopax.doc.dh_covalentbindingfeature Module#
Classes#
Class covalentbindingfeature_DocHelper |
Class Inheritance Diagram#

biopax-explorer.biopax.doc.dh_degradation Module#
Classes#
Class degradation_DocHelper |
Class Inheritance Diagram#

biopax-explorer.biopax.doc.dh_deltag Module#
Classes#
Class deltag_DocHelper |
Class Inheritance Diagram#

biopax-explorer.biopax.doc.dh_dna Module#
Classes#
Class dna_DocHelper |
Class Inheritance Diagram#

biopax-explorer.biopax.doc.dh_dnareference Module#
Classes#
Class dnareference_DocHelper |
Class Inheritance Diagram#

biopax-explorer.biopax.doc.dh_dnaregion Module#
Classes#
Class dnaregion_DocHelper |
Class Inheritance Diagram#

biopax-explorer.biopax.doc.dh_dnaregionreference Module#
Classes#
Class dnaregionreference_DocHelper |
Class Inheritance Diagram#

biopax-explorer.biopax.doc.dh_entity Module#
Classes#
Class entity_DocHelper |
Class Inheritance Diagram#

biopax-explorer.biopax.doc.dh_entityfeature Module#
Classes#
Class entityfeature_DocHelper |
Class Inheritance Diagram#

biopax-explorer.biopax.doc.dh_entityreference Module#
Classes#
Class entityreference_DocHelper |
Class Inheritance Diagram#

biopax-explorer.biopax.doc.dh_entityreferencetypevocabulary Module#
Classes#
Class entityreferencetypevocabulary_DocHelper |
Class Inheritance Diagram#

biopax-explorer.biopax.doc.dh_evidence Module#
Classes#
Class evidence_DocHelper |
Class Inheritance Diagram#

biopax-explorer.biopax.doc.dh_evidencecodevocabulary Module#
Classes#
Class evidencecodevocabulary_DocHelper |
Class Inheritance Diagram#

biopax-explorer.biopax.doc.dh_experimentalform Module#
Classes#
Class experimentalform_DocHelper |
Class Inheritance Diagram#

biopax-explorer.biopax.doc.dh_experimentalformvocabulary Module#
Classes#
Class experimentalformvocabulary_DocHelper |
Class Inheritance Diagram#

biopax-explorer.biopax.doc.dh_fragmentfeature Module#
Classes#
Class fragmentfeature_DocHelper |
Class Inheritance Diagram#

biopax-explorer.biopax.doc.dh_gene Module#
Classes#
Class gene_DocHelper |
Class Inheritance Diagram#

biopax-explorer.biopax.doc.dh_geneticinteraction Module#
Classes#
Class geneticinteraction_DocHelper |
Class Inheritance Diagram#

biopax-explorer.biopax.doc.dh_interaction Module#
Classes#
Class interaction_DocHelper |
Class Inheritance Diagram#

biopax-explorer.biopax.doc.dh_interactionvocabulary Module#
Classes#
Class interactionvocabulary_DocHelper |
Class Inheritance Diagram#

biopax-explorer.biopax.doc.dh_kprime Module#
Classes#
Class kprime_DocHelper |
Class Inheritance Diagram#

biopax-explorer.biopax.doc.dh_modificationfeature Module#
Classes#
Class modificationfeature_DocHelper |
Class Inheritance Diagram#

biopax-explorer.biopax.doc.dh_modulation Module#
Classes#
Class modulation_DocHelper |
Class Inheritance Diagram#

biopax-explorer.biopax.doc.dh_molecularinteraction Module#
Classes#
Class molecularinteraction_DocHelper |
Class Inheritance Diagram#

biopax-explorer.biopax.doc.dh_pathway Module#
Classes#
Class pathway_DocHelper |
Class Inheritance Diagram#

biopax-explorer.biopax.doc.dh_pathwaystep Module#
Classes#
Class pathwaystep_DocHelper |
Class Inheritance Diagram#

biopax-explorer.biopax.doc.dh_phenotypevocabulary Module#
Classes#
Class phenotypevocabulary_DocHelper |
Class Inheritance Diagram#

biopax-explorer.biopax.doc.dh_physicalentity Module#
Classes#
Class physicalentity_DocHelper |
Class Inheritance Diagram#

biopax-explorer.biopax.doc.dh_protein Module#
Classes#
Class protein_DocHelper |
Class Inheritance Diagram#

biopax-explorer.biopax.doc.dh_proteinreference Module#
Classes#
Class proteinreference_DocHelper |
Class Inheritance Diagram#

biopax-explorer.biopax.doc.dh_provenance Module#
Classes#
Class provenance_DocHelper |
Class Inheritance Diagram#

biopax-explorer.biopax.doc.dh_publicationxref Module#
Classes#
Class publicationxref_DocHelper |
Class Inheritance Diagram#
biopax-explorer.biopax.doc.dh_relationshiptypevocabulary Module#
Classes#
Class relationshiptypevocabulary_DocHelper |
Class Inheritance Diagram#

biopax-explorer.biopax.doc.dh_relationshipxref Module#
Classes#
Class relationshipxref_DocHelper |
Class Inheritance Diagram#
biopax-explorer.biopax.doc.dh_rna Module#
Classes#
Class rna_DocHelper |
Class Inheritance Diagram#

biopax-explorer.biopax.doc.dh_rnareference Module#
Classes#
Class rnareference_DocHelper |
Class Inheritance Diagram#

biopax-explorer.biopax.doc.dh_rnaregion Module#
Classes#
Class rnaregion_DocHelper |
Class Inheritance Diagram#

biopax-explorer.biopax.doc.dh_rnaregionreference Module#
Classes#
Class rnaregionreference_DocHelper |
Class Inheritance Diagram#

biopax-explorer.biopax.doc.dh_score Module#
Classes#
Class score_DocHelper |
Class Inheritance Diagram#

biopax-explorer.biopax.doc.dh_sequenceinterval Module#
Classes#
Class sequenceinterval_DocHelper |
Class Inheritance Diagram#

biopax-explorer.biopax.doc.dh_sequencelocation Module#
Classes#
Class sequencelocation_DocHelper |
Class Inheritance Diagram#

biopax-explorer.biopax.doc.dh_sequencemodificationvocabulary Module#
Classes#
Class sequencemodificationvocabulary_DocHelper |
Class Inheritance Diagram#

biopax-explorer.biopax.doc.dh_sequenceregionvocabulary Module#
Classes#
Class sequenceregionvocabulary_DocHelper |
Class Inheritance Diagram#

biopax-explorer.biopax.doc.dh_sequencesite Module#
Classes#
Class sequencesite_DocHelper |
Class Inheritance Diagram#

biopax-explorer.biopax.doc.dh_smallmolecule Module#
Classes#
Class smallmolecule_DocHelper |
Class Inheritance Diagram#

biopax-explorer.biopax.doc.dh_smallmoleculereference Module#
Classes#
Class smallmoleculereference_DocHelper |
Class Inheritance Diagram#

biopax-explorer.biopax.doc.dh_stoichiometry Module#
Classes#
Class stoichiometry_DocHelper |
Class Inheritance Diagram#

biopax-explorer.biopax.doc.dh_templatereaction Module#
Classes#
Class templatereaction_DocHelper |
Class Inheritance Diagram#

biopax-explorer.biopax.doc.dh_templatereactionregulation Module#
Classes#
Class templatereactionregulation_DocHelper |
Class Inheritance Diagram#

biopax-explorer.biopax.doc.dh_tissuevocabulary Module#
Classes#
Class tissuevocabulary_DocHelper |
Class Inheritance Diagram#

biopax-explorer.biopax.doc.dh_transport Module#
Classes#
Class transport_DocHelper |
Class Inheritance Diagram#

biopax-explorer.biopax.doc.dh_transportwithbiochemicalreaction Module#
Classes#
Class transportwithbiochemicalreaction_DocHelper |
Class Inheritance Diagram#

biopax-explorer.biopax.doc.dh_unificationxref Module#
Classes#
Class unificationxref_DocHelper |
Class Inheritance Diagram#
biopax-explorer.biopax.doc.dh_utilityclass Module#
Classes#
Class utilityclass_DocHelper |
Class Inheritance Diagram#

biopax-explorer.biopax.doc.dh_xref Module#
Classes#
Class xref_DocHelper |
Class Inheritance Diagram#
